Pathogenicity Calculator¶
The ClinGen Pathogenicity Calculator automates the formal reasoning process to interpret pathogenicity of variants, eliminates errors in rule application and makes it possible to automatically calculate provisional conclusions based on latest evidence. A low level API which is used in the Calculator application is available. Here are the steps to use this API to get the output (interpretations) of the variant pathogenicity reasoner.
CAVEAT: As mentioned, these are low level APIs used internally by our applications and are not meant to be distributed for widespread usage.
A. Variant Pathogenicity Reasoner API¶
A.1 IMPORTANT - Steps to do before you can use the API¶
- Ensure you have a valid account (user name and password) to use the ClinGen Pathogenicity Calculator.
- Use the GUI at http://calculator.clinicalgenome.org/site/cg-calculator to "Start a new interpretation" for your variant. You can use either CAId (ClinGen Allele Registry identifier) or an HGVS expression to start your interpretation.
- Go to the "Calculator" page and use the "Create" button to start creating new Evidence.
- Enter the "Condition" name and "Mode of Inheritance".
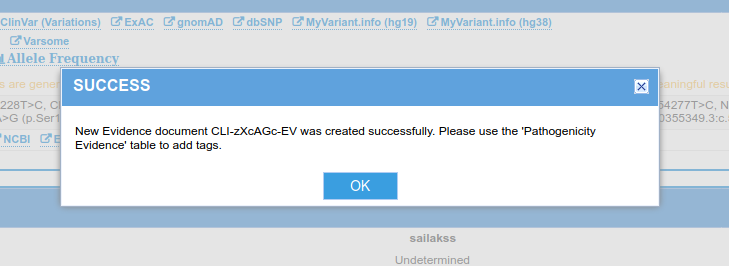
- Hit Save. When the information is saved, you will see a popup window that shows:
- The Evidence document ID will be displayed (see screenshot below for reference). This ID will be of the form: "CLI-{6}-EV" where {6} is a string with 6 alphanumeric characters. This is unique to the evidence document you created. Save this ID since this is needed to use the API below.
- Continue adding the evidence tags for different categories in the grid. Once you are done with adding all evidence, proceed to the next step to get the interpretations using the API.
A.2 Prerequisites¶
- Ruby
- Ruby Gems:
- json
- digest/sha1
- rest-client
- net/http
A.3 Using the API¶
- IMPORTANT NOTE: Make sure you added all evidence using the UI and also ensure you have the Evidence document ID from section A.1 above.
- The steps given below can be run on an
irb(Interactive Ruby) session. - To start this session, open the Terminal in your computer and issue the command
irb. - Wherever it appears, replace "{YOUR_LOGIN}" with your login name and "{YOUR_PASSWORD}" with your password used to login to the Pathogenicity Calculator UI
Similarly, replace "{EVIDENCE_DOC_ID_FROM_UI}" with the evidence doc ID you copied over from the UI (in A.1).
require 'json'
require 'digest/sha1'
require 'rest-client'
require 'net/http'
CALCHOST = 'http://calculator.clinicalgenome.org'
# Function to build the authentication token for your login credentials and group in pathogenicity calculator application
def buildAuthToken(rsrcURI, gbLogin, userPassword, gbTime = Time.now.to_i)
gbToken = getgbToken rsrcURI, gbLogin, userPassword, gbTime
return "gbLogin=#{gbLogin}&gbTime=#{gbTime}&gbToken=#{gbToken}".strip
end
# Function to create the authentication token - SHA1 hexdigest
def getgbToken(rsrcURI, gbLogin, userPassword, gbTime = Time.now.to_i)
credential = Digest::SHA1.hexdigest gbLogin + userPassword
Digest::SHA1.hexdigest rsrcURI + credential + gbTime.to_s
end
# Provide details such as login, group name
grp = "{YOUR_LOGIN}"
kb = "{YOUR_LOGIN}"
coll = "Evidence"
kbdoc = "{EVIDENCE_DOC_ID_FROM_UI}"
# URL of evidence document
kbDocUrl = "#{CALCHOST}/REST/v1/grp/#{grp}/kb/#{kb}/coll/#{coll}/doc/#{kbdoc}"
# Transformation document that maps evidence code to Pathogenicity and Strength
kbTransformUrl = "#{CALCHOST}/REST/v1/grp/pcalc_resources/kb/pcalc_resources/trRulesDoc/acmgTransform"
# This is the ACMG 2015 guideline document
rulesDocUrl = "#{CALCHOST}/REST/v1/grp/pcalc_resources/kb/pcalc_resources/coll/GuidelineRulesMetaRules/doc/ACMG2015-Guidelines"
# Genboree job to run the reasoning engine for variant pathogenicity interpretation
jobUrl = "#{CALCHOST}/REST/v1/genboree/tool/reasonerV2a1/job?"
uri = "#{jobUrl}#{buildAuthToken(jobUrl,"{YOUR_LOGIN}","{YOUR_PASSWORD}")}"
#puts uri
# Prepare the job specifications
jobConf = {
"inputs" => [kbDocUrl, kbTransformUrl],
"outputs" => [],
"settings" => {
"rulesDoc" => rulesDocUrl
},
"context" => {}
}
# Run the calculator job to get the interpretations in JSON format - API PUT Request
response = RestClient.put uri, jobConf.to_json , {:content_type => :json}
A.4 Response from the API¶
- The JSON API response contains the reasoner output, which looks similar to this:
"data": {
"Reasoner output": {
"properties": {
"Status": {
"properties": {
"Message": {
"value": "ok"
} ,
"Name": {
"value": "ok"
}
} ,
"value": "ok"
} ,
"FinalCall": {
"properties": {
"Text": {
"value": "Two assertions: Pathogenic and Uncertain Significance - Conflicting Evidence were made. The final call in this case is Uncertain Significance - Conflicting Evidence."
}
} ,
"value": "Uncertain Significance - Conflicting Evidence"
} ,